Activity-Based Proteome Profiling
Activity-based proteome profiling (ABPP) is an attractive and powerful chemical biological technique that accelerates most stages of the drug discovery process.
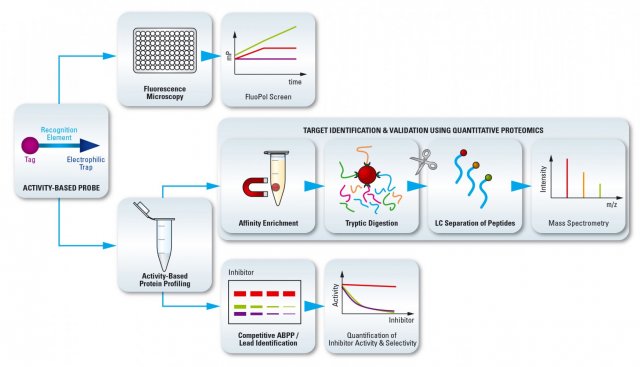
ABPP allows us to study the function of proteins and their perturbation by small molecules in their native cellular context. ABPP It uses probes to report on protein’s activity in cells, tissue or animals. ABPP will only visualize active proteins and takes all post-translational modifications (PTMs) into account and by this virtue is complementary to other techniques that detect messenger RNA or proteins (that is, in situ hybridization and immunohistology, respectively). ABPP makes use of organic molecules, termed activity-based probes (ABPs) to label the active site of a protein. ABPs are compounds that covalently and irreversibly inhibit enzymes and that are equipped with a tag (fluorophore, biotin, bioorthogonal tag) through which the target enzyme, or enzyme family, is visualized in living systems by fluorescence microscopy, or enriched to enable identification and characterization using chemical proteomics methodology by mass spectrometers.
ABPP offers a versatile toolbox of different experimental modalities to create new opportunities to identify novel targets, to discover leads, to inform on lead selectivity across the whole proteome and to establish target engagement in vitro, in situ, in tissues and intact animals. In comparative ABPP two biological samples (for instance, diseased vs healthy control) are interrogated with ABPs. Differences in enzyme activity are monitored and identified with various ABPs. Comparative ABPP has emerged as a powerful tool both for the discovery of targets and validation of drug-target interaction in live cells, tissue lysates, and animals and for revealing the presence and involvement of such active proteins in a given biological process under investigation. In competitive ABPP a small molecule (for instance from a focused library, or a hit from a screening campaign) is pre-incubated with a biological sample and residual enzyme activities are subsequently monitored with an ABP. Activity and selectivity of the small molecules is easily visualized in a complex proteome across the complete protein family (lead identification).
Competitive ABPP can also be used to determine target engagement in cellular and living systems. In both experimental set ups, two different type of probes can be used. Broad-spectrum ABPs target a whole (or to a large extent) family of proteins, whereas tailor-made ABPs are designed to target a specific protein of interest. The latter type of probe can also be used to validate the target in different therapeutic animal models and serve as a biomarker for target engagement in clinical trials. In case the ABPs fall short and does not work due to a lack of bioavailability or enzyme specificity, two-step ABPs can be applied. Two-step ABPs do not constitute a reporter tag, but instead carry a small ligation handle, which can be conjugated to a biotin or fluorescent tag via bio-orthogonal ligation chemistry, only after the ABP has covalently reacted with the target of interest. These combined ABPP technologies provide a highly attractive platform, both to discern aberrant enzyme functioning in physiological processes, and to identify compounds able to correct for this. Furthermore, ABPP may guide the design, optimization and selection of drug candidates. NL Openscreen has wide experience the design, synthesis and use of activity-based probes for a wide range of protein families. We have made various contributions to the ABPP field and have available a comprehensive set of unique ABP tools targeting a variety of enzymes and enzyme families. We have, for example, recently applied ABPP to discover the off-target profile of the fatal drug BIA 10-2474, thereby providing a potential explanation for the severe side effects observed in a phase I clinical trial in France in 2016 (Esbroeck et al., Science, 2017).